Smrutisanjita Behera , Ph.D.
Scientist
Infectious Diseases and Immunology

Research Interest
Investigating Vesicle-Mediated Signaling in Rice–Xoo Interactions
Bacterial blight of rice, caused by the Gram-negative bacterium Xanthomonas oryzae pv. oryzae (Xoo), is a devastating disease affecting rice cultivation across Asia. Our research group is focused on deciphering the early signaling events that occur during the interaction between rice and Xoo, with particular attention to newly identified molecular components involved in this process.
Outer membrane vesicles (OMVs) secreted by Xoo play a pivotal role in environmental sensing and host-pathogen communication. Certain Xanthomonas species can trigger immune responses in plants, while rice also responds to pathogen invasion by releasing extracellular vesicles (EVs). These plant-derived EVs are emerging as key players in plant immunity. However, the precise mechanisms by which these vesicles mediate interspecies communication remain largely unknown.
Our current research aims to address the following aspects of vesicle-mediated plant–microbe interactions:
- How rice perceives OMVs produced by Xoo
- The contribution of Xoo OMVs to pathogenicity
- Characterization of rice-derived extracellular vesicles
- Functional roles of rice EVs during pathogen attack
- The interplay of second messengers such as reactive oxygen species (ROS) and calcium ions (Ca²⁺) in signalling
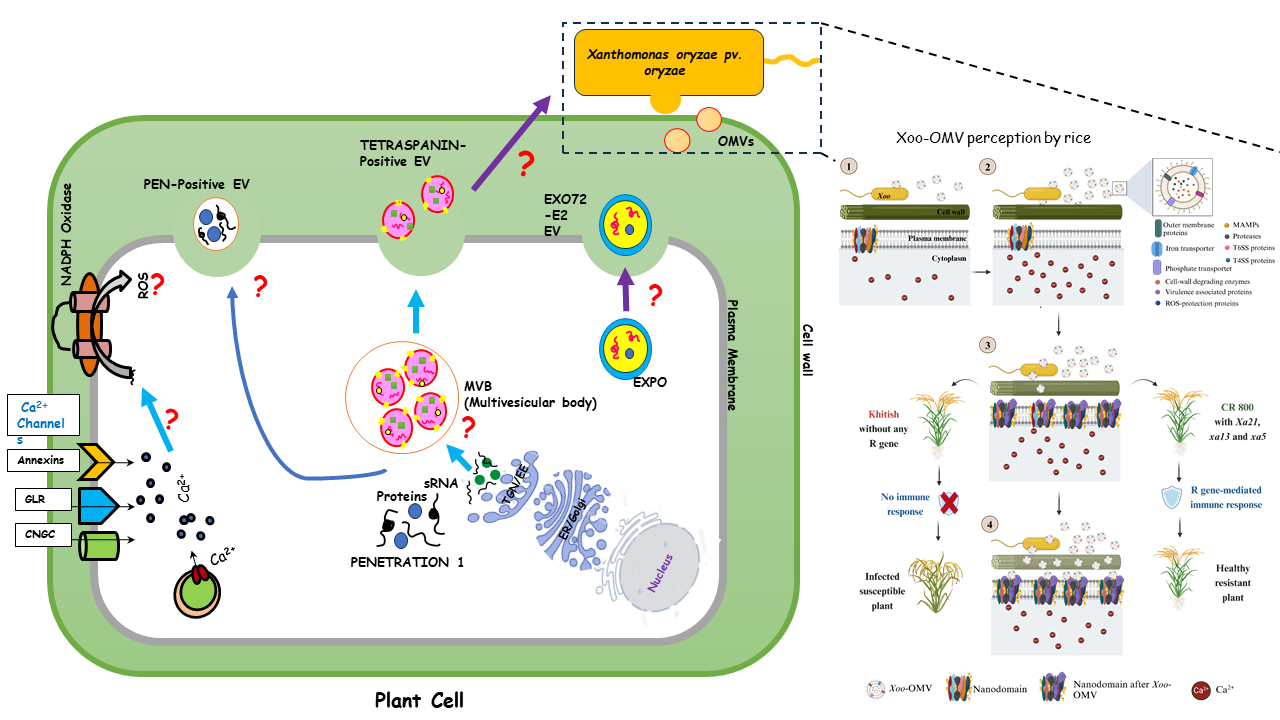
Through this work, we aim to uncover novel insights into the molecular dialogue between rice and Xoo, with the goal of advancing our understanding of plant immune responses and pathogen strategies.
Credentials
- DST-INSPIRE faculty, Indian Institute of Chemical Biology, Kolkata, India (2016-2021)
- Postdoctoral Research, University of Milan, Italy (2014-2015)
- Research Associate, University of Münster, Germany (2013-2014)
- PhD, University of Münster, Germany (2008-2013)
- MSc, Jawaharlal Nehru University, New Delhi, School of Life sciences (2005-2007)
Honours & Awards
- DST-INSPIRE faculty award, 2016
- Post doctoral fellowship from Programma ‘Futuro in Ricerca 2010’, Italy within the scientific area of Biological Sciences
- International fellowship from CEDAD-IMPRS International Graduate School (2008-2011)
- Fellowship from Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR), Italy (2007-2008)
- Junior Research Fellowship and Eligibility for Lectureship conducted by CSIR-UGC (2007)
Patents & Publications
PUBLICATIONS:
- Dutta S, Kothari S, Singh D, Ghosh S, Sarangi N, Behera S, Prajapati S, Sinha P K, Prusty A, Tripathy S. Novel oceanic cyanobacterium isolated from Bangaram island with profound acid neutralizing ability is proposed as Leptolyngbya iicbica sp. nov. strain LK. Molecular Phylogenetic Evolution. 2024 (197) 108092. doi: 10.1016/j.ympev.2024.108092
- Mittal D, Gautam J K, Varma M, Laie A, Mishra S, Behera S, Vadassery J. External jasmonic acid isoleucine mediates amplification of plant elicitor peptide receptor (PEPR) and jasmonate-based immune signalling. Plant cell and environment. 2024 (4)1397-1415. doi:org/10.1111/pce.14812
- Steinhorst L, He G, Moore KL, Schültke S, Schmitz Thom1 I, Cao Y, Hashimoto K, Andrés Z, Piepenburg K, Ragel P, Behera S et al. A Ca2+-sensor switch for tolerance to elevated salt stress in Arabidopsis. Developmental cell. 2022 (17) 2081-2094. doi: 10.1016/j.devcel.2022.08.001
- Mukherjee M, Geeta A, Ghosh S, Prusty A, Dutta S, Sarangi AN, Behera S, Adhikary SP, Tripathy S. Genome Analysis Coupled With Transcriptomics Reveals the Reduced Fitness of a Hot Spring Cyanobacterium Mastigocladus laminosus UU774 Under Exogenous Nitrogen Supplement. Front Microbiol. 2022 (13) 909289. doi: 10.3389/fmicb.2022.909289
- Yuan P, Jewell JB, Behera S, Tanaka K, Poovaiah BW. Distinct Molecular Pattern-Induced Calcium Signatures Lead to Different Downstream Transcriptional Regulations via AtSR1/CAMTA3. International journal of molecular sciences. 2020 (21) 8163. doi: org/10.3390/ijms21218163
- Hazak O, Mamon E, Lavy M, Stenberg H, Behera S, Schmitz-Thom I, Bloch D, Dementive O, Gutman I, Danziger T, Schwarz N, Abuzeineh A, Mockaitis K, Estelle M, Hirsch J, Kudla J, Yalovsky S. A novel Ca2+-binding protein that can rapidly transduce auxin responses during root growth. PLOS Biology. 2019 (7) e3000085. doi: org/10.1371/journal.pbio.3000085
- Behera S, Zhaolong X, Luoni L, Bonza MC, Doccula FG, De Michelisa MI, Morris RJ, Schwarzländere M, Costa A. Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell. 2018 (11) 2704-2719. doi: 10.1105/tpc.18.00655
- Teardo E, Carraretto L, Wagner S, Formentin E, Behera S, De Bortoli S, Larosa V, Fuchs P, Lo Schiavo F, Raffaello A, Rizzuto R, Costa A, Schwarzländer A, Szabò I. Physiological characterization of a plant mitochondrial calcium uniporter in vitro and in vivo. Plant Physiology 2017 (173) 1355-1370. doi: 10.1104/pp.16.01359
- Hussain J, Chen J, Locato V, Sabetta W, Behera S, Cimini S, Griggio F, Martínez-Jaime S, Graf A, Bouneb M, Pachaiappan R, Fincato P, Blanco E, Costa A, De Gara L, Bellin D, De Pinto MC, Vandelle E: Constitutive cyclic GMP accumulation in Arabidopsis thaliana compromises systemic acquired resistance induced by an avirulent pathogen by modulating local signals. Scientific Reports 2017 (7) 46593. doi: 10.1038/srep36423.
- Yu J, Meng Z, Liang W, Behera S, Kudla J, Tucker MR, Luo Z, Chen M, Xu D, Zhao G, Wang J, Zhang S, Kim YJ, Zhang D. A Rice Ca2+ Binding Protein Is Required for Tapetum Function and Pollen Formation. Plant Physiology. 2016 Nov 23;172(3):1772-1786. doi: 10.1104/pp.16.01261
- Behera S, Long Y, Schmitz-Thom I, Wang X-P, Zhang C, Li H, Steinhorst L, Manishankar P, Ren X-L, Offenborn J N, Wu W H, Kudla J, Yi W: Two spatially and temporally distinct Ca2+ signals convey Arabidopsis thaliana responses to K+ deficiency. New Phytologist. 2017 (213) 739 -750. doi: 10.1111/nph.14145
- Loro G, Wagner S, Doccula FG, Behera S, Stefan S, Kudla J, Schwarzländer M, Costa A, Zottini M. Chloroplast-specific in vivo Ca2+ imaging using Yellow Cameleon fluorescent protein sensors reveals organelle-autonomous Ca2+ signatures in the stroma. Plant Physiology. 2016 Aug 1;171(4):2317-2330. doi: 10.1104/pp.16.00652
- Wagner S, Behera S, De Bortoli S, Logan D C, Fuchs P, Carraretto L, Teardo E, Cendron L, Nietzel T, Füßl M, Doccula F G, Navazio L, Fricker M D, Van Aken O, Finkemeier I, Meyer A J, Szabò I, Costa A, Schwarzländer M. The EF-Hand Ca2+ Binding Protein MICU Choreographs Mitochondrial Ca2+ Dynamics in Arabidopsis. The Plant Cell. 2015 (27) 3190-3212. doi: 10.1105/tpc.15.00509
- Behera S, Wang N, Zhang C, Schmitz‐Thom I, Strohkamp S, Schültke S, Hashimoto K, Xiong L, Kudla J. Analyses of Ca2+ dynamics using a ubiquitin-10 promoter-driven Yellow Cameleon 3.6 indicator reveal reliable transgene expression and differences in cytoplasmic Ca2+ responses in Arabidopsis and rice (Oryza sativa) roots. New Phytologist. 2015 (206) 751-760. doi: 10.1111/nph.13250
- Teardo E, Carraretto L, De Bortoli S, Costa A, Behera S, Wagner R, Lo Schiavo F, Formentin E, Szabo I. Alternative splicing-Mediated targeting of the arabidopsis GLUTAMATE RECEPTOR 3.5 to mitochondria affects organelle morphology. Plant Physiology 2015 (167) 216-227. doi: 10.1104/pp.114.242602
- Bonza MC, Loro G, Behera S, Wong A, Kudla J, Costa A: Analyses of Ca2+ Accumulation and Dynamics in the Endoplasmic Reticulum of Arabidopsis Root Cells Using a Genetically Encoded Cameleon Sensor. Plant Physiology. 2013 (163) 1230-1241. doi: 10.1104/pp.113.226050
- Costa A, Drago I, Behera S, Zottini M, Pizzo P, Schroeder J I, Pozzan T, Lo Schiavo F: H2O2 in plant peroxisomes: An in vivo analysis uncovers a Ca2+-dependent scavenging system. The Plant Journal. 2010 (62) 760-772. doi: 10.1111/j.1365-313X.2010.04190.x
PROTOCOLS:
- Doccula FG, Luoni L, Behera S, Bonza MC, Costa A. In vivo analysis of calcium levels and glutathione redox status in Arabidopsis epidermal leaf cells infected with hypersensitive response-inducing Bacteria Pseudomonas syringe pv. tomato AvrB (PstAvrB). 2018. Methods in Molecular Biology. 1743:125-141. ISSN: 1064-3745
- Behera S, Krebs M, Loro G, Schumacher K, Costa A, Kudla J: Ca2+ imaging in plants using genetically encoded Yellow Cameleon Ca indicators. Cold Spring Harbor Protocols 08/2013; 2013(8). DOI:10.1101/pdb.top066183 ISBN:978-1-936113-58-3
- Behera S, Kudla J: High-Resolution Imaging of Cytoplasmic Ca2+ Dynamics in Arabidopsis Roots. Cold Spring Harbor Protocols 07/2013; 2013(7). DOI:10.1101/pdb.prot073023 ISBN:978-1-936113-58-3
- Behera S, Kudla J: Live Cell Imaging of Cytoplasmic Ca2+ Dynamics in Arabidopsis Guard Cells. Cold Spring Harbor Protocols 07/2013; 2013(7). DOI:10.1101/pdb.prot072983 ISBN:978-1-936113-58-3
